Faculty
|

|
Please click here for more information.
|
Senior Scientists
|

|
Yibing Wu (C.V.)
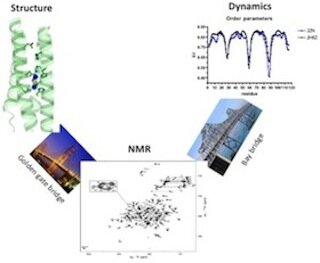
My research in general focuses on structure and dynamic study of protein using multidimensional NMR spectroscopy. At different Zn concentrations, I have recently solved two Diiron structures, which help us make new designs to stabilize the structure and optimize the active site. Structural study on AM2 protein is another ongoing project. Current focus is to identify good inhibitors and obtain high-resolution solution structure of M2 S31N in presence of drug to investigate the detail binding sites.
|
Postdocs
|

|
Jenny Hu
I'm working on a chromosomally encoded protein of the plant pathogen Agrobacterium tumefaciens that belongs to a family of periplasmic sugar binding protein - ChvE, which is homologous to ribose-binding protein (RBP) and galactose/glucose-binding protein (GGBP) of Escherichia coli. ChvE is proposed to be involved in the virulence of A. tumefaciens and sugar transport. When it binds sugars, it changes its conformation and binds to a signal transduction protein that is a two-component system involved in virulence. In the sugar-bound state, ChvE can pass the sugar to a transporter, allowing the bacterium to use the sugar for growth. We are interested in understanding the mechanisms of sugar recognition and computational design mutants intended to bind different sugars. Currently, we are working on the crystallization of ChvE.
|

|
Hyunil Jo (C.V.)
My research area spans from Medicinal Chemistry to Protein Engineering. I have been working on developments of antimalarial, anti-HIV, Integrin inhibitors, Splicing corrector for SMA (Spinal Muscular Atrophy) treatment, and M2 proton channel inhibitor. For protein engineering, protein structure restriction by side chain modification, installation of IR and UV-resonance Raman probe have been intensively studied.
|

|
Nate Joh (C.V.) ()
Transmembrane transport, central to many vital cell functions including nutrient uptake, signaling and drug efflux, is mediated by various transporters. To learn the transporter function and mechanism, I take both "top-down" and "bottom-up" approaches.
In a top-down approach where I learn by examining the natural system, proton conduction by matrix protein M2 from influenza A virus is investigated, as well as its neutralization by anti-M2 antibodies, using various biophysical methods including crystallography.
Alternatively, in a bottom-up approach, I apply the first principles to generate the transport functionality through engineering the alternate-access mechanism using de-novo protein design method.
|

|
Michelle McCully (C.V.)
|

|
Gözde Ulas
My research involves the design of small peptides in order to modulate activity of target protein complexes. By stabilizing key intermediates of target protein function, we aim to capture transient stages of proteins of interest. One intermediary stage that we would like to stabilize is the intermediary fusion stage of HIV-1 gp41 envelope glycoprotein.
|

|
Jun Wang (C.V.)
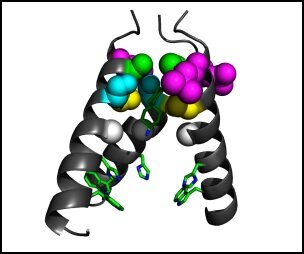
I am focusing on structure based drug design of influenza A virus M2 channel inhibitors. As most of the currently circulating human influenza A virus carrying amantadine resistant A/M2 mutations, there is a great need for novel anti-influenza drugs. We approach this problem by iterative cycles of design, synthesis, assay and structure-activity correlation (SAR) study. In addition, structural characterization of the mode of drug binding to A/M2 by NMR, X-ray etc offers invaluable information in guiding drug design.
|
Grduate Students
|

|
Gabriel Gonzalez
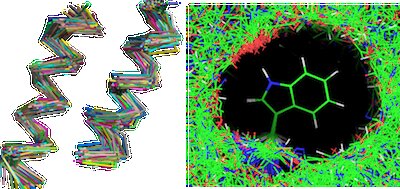
I design proteins computationally by data-mining the Protein Data Bank (PDB). I've authored a high-performance structural search algorithm that can be used to automatically generate backbones, suggest residues, and validate designs, among many other features.
|

|
Brett Hannigan
|

|
Kathleen Molnar
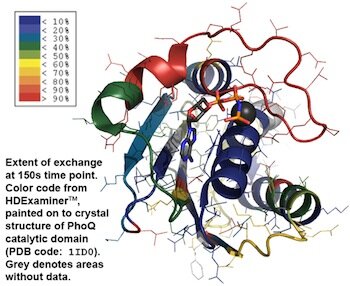
Mass spectrometry is critical to many quantitative biophysical techniques. One such technique, hydrogen/deuterium exchange is used to measure the dynamics of proteins in vitro. Pictured below are the exchange dynamics of the catalytic domain of PhoQ. My research focuses this bacterial two-component system PhoQ/P and determining the protein dynamics during signaling.
|

|
Jessica Thomaston
My primary interests are membrane protein structure and function. Currently, the focus of my efforts is a structural study of the AM2 and BM2 proton channels of influenza. Influenza AM2 was a drug target before viral resistance developed in the past decade, and the BM2 channel has no known ligands. I use lipidic cubic phase (LCP) techniques in an attempt to crystallize these transmembrane channels in a physiologically relevant environment.
|

|
Shaoqing Zhang
My current projects focus on the relationship between the sequence and conformation of transmembrane helices using structural bioinformatic approaches. I am also working on the structural and functional analysis of transmembrane proteins in signal transduction, specifically in the Tpo/TpoR system.
|
Staff
|

|
Lili Li
Administrative Assistant for the lab
|

|
Bruk Mensa
Recent concerns in antibacterial-resistant bacteria strains have sparked in-depth research into AMPs as a potential venue for future drug-development. Although it has been shown that these AMPs show appreciable bactericidal activity against a large number of pathogens, concerns about the clinical applications of these peptides has led to the development of synthetic analogues that make use of the same principles of AMPs (charge and amphiphilicity) with the added advantages of proteolytic stability and mass-scale production potential. These mimics include aryl-amide and phenylene-ethynylene compounds and have been shown to be remarkably effective against a wide range of bacteria with no apparent resistance development. In vitro assays and MD simulations suggest that these compounds may cause perturbations in the membrane and low-level leakage of vesicle contents. However, the nature of their interaction with bacteria, and their variable activities and selectivities with different R-group substitutions remains largely uncharacterized. I am currently using 2 model organisms, the gram-negative bacterium E. coli and the gram-positive bacterium S. aureus, to study the transcriptional response of bacteria to sub-lethal concentrations of these synthetic mimics. This and other biochemical in vivo assays can potentially elucidate bacterial sensing pathways, potential resistance mechanisms, and modes of action for these compounds that explain our primary observations, and explain the similarities and differences to natural AMPs.
|





















